Multiple sequence alignment with top benchmark scores scalable to thousands of sequences. Generates replicate alignments, enabling assessment of downstream analyses such as trees and predicted structures.
Muscle is widely-used software for making multiple alignments of biological sequences.
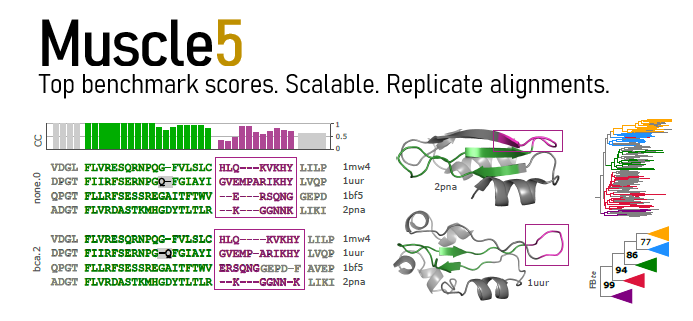
Muscle achieves highest scores on Balibase, Bralibase and Balifam benchmark tests and scales to thousands of sequences or structures on a commodity desktop computer.
Muscle supports generating an ensemble of alternative alignments with the same high accuracy obtained with default parameters. By comparing downstream predictions from different alignments, such as trees, a biologist can evaluation the robustness of conclusions against alignment variation caused by ambiguities and errors.
Multiple structure alignment
Structure alignment (“Muscle-3D”) is supported as well as conventional amino acid sequence alignment. Input for structure alignment is a “mega” file generated by the pdb2mega command of reseek (https://github.com/rcedgar/reseek).
# for up to ~100 structures reseek -pdb2mega STRUCTS -output structs.mega muscle -align structs.mega -output structs.afa # for up to ~10,000 structures reseek -convert STRUCTS -bca structs.bca reseek -pdb2mega structs.bca -output structs.mega reseek -distmx structs.bca -output structs.distmx muscle -super7 structs.mega -distmxin structs.distmx -reseek -output structs.afa
Downloads and installation
Binary files are self-contained, no dependencies. To install, download the binary and make sure the execute bit is set.
https://github.com/rcedgar/muscle/releases
Documentation
Building MUSCLE from source
https://github.com/rcedgar/muscle/wiki/Building-MUSCLE
References
Edgar RC., Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nature Communications 13.1 (2022): 6968.
https://www.nature.com/articles/s41467-022-34630-w.pdf
Edgar RC. and Tolstoy I., Muscle-3D: scalable multiple protein structure alignment (2024) BioRxiv.
